Overview
bioSDP is a Matlab Toolbox for the analysis of uncertain biochemical networks via semidefinite programming. Its main features are:
- Computation of uncertainty ranges for stationary states under parametric uncertainty;
- Computing the set of consistent parameters for uncertain measurement data.
- Computing a parameter set for guaranteed robustness of steady state concentrations.
Download & Installation
All released versions of the software are available for download on the Sourceforge download page of the project. The latest release is 0.3, published May 29, 2012.
To get the latest developer's version of the toolbox, you can access the Subversion repository, for example via the command
svn export http://biosdp.svn.sf.net/svnroot/biosdp/trunk biosdp
Software requirements for bioSDP are Matlab (Release 2007b or later), and the SeDuMi toolbox version 1.1 or later. To install, add the 'toolbox' directory of the bioSDP package to the Matlab path. The best way to achieve this is to put a lign like
addpath(genpath("/path/to/toolbox"))
into your Matlab startup script.
Usage
In order to get started, we recommended reading the online tutorial. There, the basic usage of bioSDP is shown with small examples.
The bioSDP Toolbox is also documented within the Matlab help system. Type
>> help biosdpon the Matlab command prompt in order to get an overview.
Before starting your own projects, it may also be helpful to have a look at the example applications which are provided in the examples directory of the bioSDP package.
Literature
The following publications describe the algorithms behind the bioSDP Toolbox and contain detailed descriptions of application examples.
- Waldherr, S.; Findeisen, R. & Allgöwer, F.: Global Sensitivity Analysis of Biochemical Reaction Networks via Semidefinite Programming. Proceedings of the 17th IFAC World Congress, Seoul, Korea, pages 9701-06, 2008.
- Hasenauer, J.; Rumschinski, P.; Waldherr, S.; Borchers, S.; Allgöwer, F. & Findeisen, R.: Guaranteed steady state bounds for uncertain (bio-)chemical processes using infeasibility certificates. Journal of Process Control 20:1076-1083, 2010.
- Hasenauer, J.; Waldherr, S.; Wagner, K. & Allgöwer, F.: Parameter Identification, Experimental Design and Model Falsification for Biological Network Models Using Semidefinite Programming. IET Systems Biology 4:119-130, 2010.
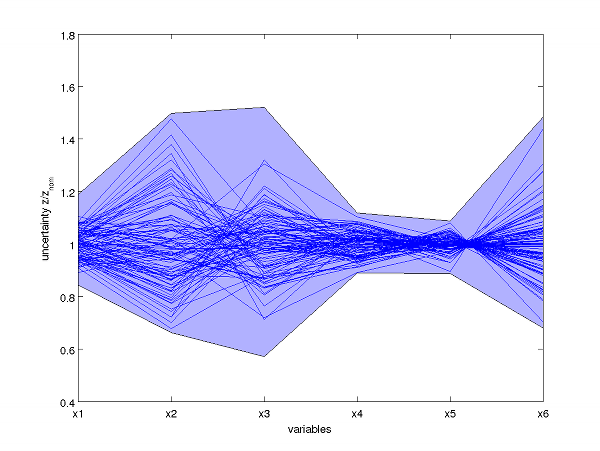
Screenshots
Visualization of steady state uncertainty ranges in parallel coordinates.
The light blue area describes upper and lower bounds on individual variables computed via semidefinite programming.
The blue lines correspond to feasible samples and are used to illustrate the tightness of the approximation.
Computation of feasible parameter set for uncertain measurement data.
The light blue area is an outer bound on the set of feasible parameters computed via semidefinite programming.
The red dots are samples of feasible parameters.
